Automating TAP queries
TOPCAT is a great tool – in particular, for prototyping and ad-hoc analyses, it's hard to beat. But say you've come up with this great TAP query, and you want to preserve it, perhaps generate plots, then maybe fix them when you (or referees) have spotted problems.
Then it's time to get acquainted with TOPCAT's command line sister STILTS. This tool lets you do most of what TOPCAT can, but without user intervention. Since the command lines usually take a bit of twiddling I usually wrap stilts calls with GNU make, so I just need to run something like make fig3.pdf or make fig3.png and make figures out what to do, potentially starting with a query. Call it workflow preservation if you want.
How does it work? Well, of course with a makefile. In that, you'll first want to define some variables that allow more concise rules later. Here's how I usually start one:
STILTS?=stilts
# VOTables are the results of remote queries, so don't wantonly throw
# them away
.PRECIOUS: %.vot
# in this particular application, it helps to have this global
HEALPIX_ORDER=6
# A macro that contains common stuff for stilts TAP query -- essentially,
# just add adql=
TAPQUERY=$(STILTS) tapquery \
tapurl='http://dc.g-vo.org/tap' \
maxrec=200000000 \
omode=out \
out=$@ \
ofmt=vot \
executionduration=14400
# A sample plot macro. Here, we do a healpix plot of some order. Also
# add value_1=<column to plot>
HEALPIXPLOT=$(STILTS) plot2sky \
auxmap=inferno \
auxlabel='$*'\
auxvisible=true \
legend=false \
omode=out \
out=$@ \
projection=aitoff \
sex=false \
layer_1=healpix \
datalevel_1=$(HEALPIX_ORDER) \
datasys_1=equatorial \
viewsys_1=equatorial \
degrade_1=0 \
transparency_1=0 \
healpix_1=hpx \
in_1=$< \
ifmt_1=votable \
istream_1=true \
For the somewhat scary STILS command lines, see the STILTS documentation or just use your intution (which mostly should give you the right idea what something is for).
The next step is to define some pattern rules; these are a (in the syntax here, GNU) make feature that let you say „to make a file matching the destination pattern when you have one with the source pattern, use the following commands”. You can use a number of variables in the rules, in particular $@ (the current target) and $< (the first prerequisite). These are already used in the definitions of TAPQUERY and HEALPIXPLOT above, so they don't necessarily turn up here:
# healpix plots from VOTables; these will plot obs
%.png: %.vot
$(HEALPIXPLOT) \
value_1=obs \
ofmt=png \
xpix=600 ypix=380
%.pdf: %.vot
$(HEALPIXPLOT) \
value_1=obs \
ofmt=pdf
# turn SQL statements into VOTables using TAP
%.vot: %.sql
$(TAPQUERY) \
adql="`cat $<`"
Careful with cut and paste: The leading whitespace here must be a Tab in rules, not just some blanks (this is probably the single most annoying feature of make. You'll get used to it.)
What can you do with it? Well, for instance you can write an ADQL query into a file density.sql; say:
SELECT count(*) as obs, -- "6" in the next line must match HEALPIX_ORDER in the Makefile ivo_healpix_index (6, alphaFloat, deltaFloat) AS hpx FROM ppmx.data GROUP BY hpx
And with this, you can say:
make density.pdf
and get a nice PDF with the plot resulting from that query. Had you just said make density.vot, make would just have executed the query and left the VOTable, e.g., for investigation with TOPCAT, and if you were to type make density.png, you'd get a nice PNG without querying the service again. Like this:
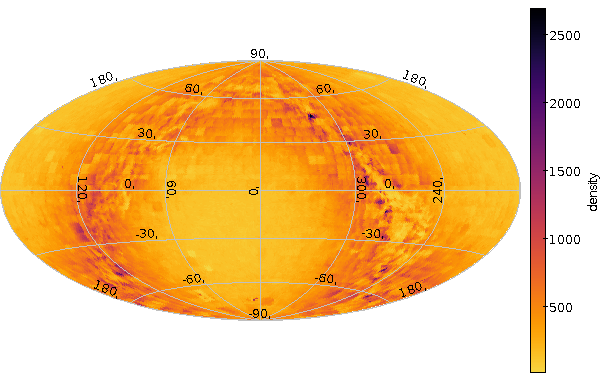
Unless of course you changed the SQL in the meantime, in which case make would figure out it had to go back to the service.
In particular for the plots you'll often have to override the defaults. Make is smart enough to figure this out. For instance, you could have two files like this:
$ cat pm_histogram.sql
SELECT
round(pmtot/10)*10 as bin, count(*) as n
FROM (
SELECT sqrt(pmra*pmra+pmde*pmde)*3.6e6 as pmtot
FROM hsoy.main) AS q
group by bin
$ cat pm_histogram_cleaned.vot
SELECT
round(pmtot/10)*10 as bin,
count(*) as n
FROM (
SELECT sqrt(pmra*pmra+pmde*pmde)*3.6e6 as pmtot
FROM hsoy.main
WHERE no_sc IS NULL) AS q
group by bin
(these were used to analyse the overall proper motions distributions in HSOY properties; note that each of these will run about 30 minutes or so, so better adapt them to what's actually interesting to you before trying this).
No special handling in terms of queries is necessary for these, but the plot needs to be hand-crafted:
pm_histograms.png: pm_histogram.vot pm_histogram_cleaned.vot
$(STILTS) plot2plane legend=false \
omode=out ofmt=png out=$@ \
title="All-sky" \
xpix=800 ypix=600 \
ylog=True xlog=True\
xlabel="PM bin [mas/yr, bin size=10]" \
xmax=4000 \
layer1=mark \
color1=blue \
in1=pm_histogram.vot \
x1=bin \
y1=n \
layer2=mark \
in2=pm_histogram_cleaned.vot \
x2=bin \
y2=n
– that way, even if you go back to the stuff six months later, you can still figure out what you queried (the files are still there) and what you did then.
A makefile to play with (and safe from cut-and-paste problems) is available from Makefile_tapsample (rename to Makefile to reproduce the examples).

![[RSS]](../theme/image/rss.png)