HTTPS in DaCHS
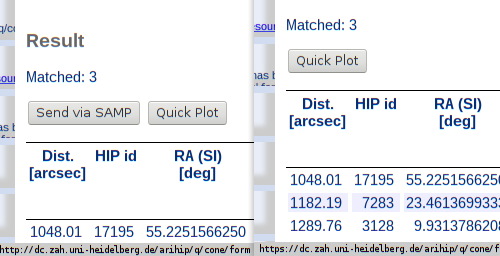
Another little aspect of HTTPS support in DaCHS: In the web interface, the webSAMP button must disappear in pages served through HTTPS: it simply wouldn't work.
(Warning: No astronomy-relevant content at all this time).
I can't say I'm a big fan of the mighty push towards HTTPS that's going on right now – as I'm arguing in the updated operator's guide it doesn't do people's privacy a lot of good (compared to, say, pushing for browsers to not execute Javascript by default or have DNSSEC widely deployed), but it's a fairly substantial operational liability. With HTTPS, operators have to deal with cryptographic material, regularly update their certificates, restart their services in time and assemble the whole thing correctly (don't get me started about proxying, SNI, and all those horrors). Users, on the other hand, have to keep their CA certificates in order, in particular when they do programmatic VO access, where the browser vendors, their employers and who knows who else doesn't do it for them. Pop quiz: How would you install a new CA certificate on your box? And will your default browser see it?
But on the other hand, there are some scenarios in which HTTPS makes sense, and I can remotely fantasise that some of those may even be relevant to the VO. And people have been asking for HTTPS in DaCHS a number of times, at times even because their administrations urged them to switch. So, here it is, hopefully. Turning it on is reasonably easy when you use Letsencrypt (which in particular entails having ports 80 and 443); the section on Letencrypt in the operator's guide tells what to do. In particular don't forget the cron job, because without it, things would break after three months (when the initial certificate expires).
Things get difficult after that. For one, if your box is known under several names (our data center, for instance, can be reached as any of dc.g-vo.org, vo.uni-hd.de, and dc.zah.uni-heidelberg.de; this of course also includes things like www.example.org and example.org), you'll now have to tell DaCHS about it in the new [web]alternateHostnames configuration item; for instance, we have:
[web] serverURL: http://dc.zah.uni-heidelberg.de alternateHostnames:dc.g-vo.org, vo.uni-hd.de
in our /etc/gavo.rc.
And then the Registry has to know you have https. There's actually no convention for that in the VO yet. But since I'd really like to have at least fallback interfaces with plain HTTP, we'll have to come up with something. For now, my plan is to have the alternative protocol (i.e., HTTPS for sites that have an HTTP-serverURL and vice versa) using the brand-new VOResource 1.1 mirrorURLs (in RegTAP 1.1, they are in the mirror_url column rr.interface). To make DaCHS declare the alternate URLs, set [web]registerAlternative to True.
Another change I've introduced for HTTPS is that the default HTML template for the form renderer (i.e., the one people use who come with a browser) now suppresses the SAMP button if the request came in through HTTPS; that's because WebSAMP doesn't work with HTTPS and probably never will – at least I can't see a way to make it happen without totally wrecking what security guarantees HTTPS gives.
All this doesn't yet cater for the case when you use a reverse proxy to terminate HTTPS. If you are in that situation, please talk to me so we can figure out a sane way for you explain to DaCHS what to tell the Registry.
Anyway, if you want to try things out, just switch to the beta repostitory and upgrade. Feedback is highly welcome.
Oh, and if you're a client developer: Our data center is now reachable through HTTPS (at https://dc.g-vo.org), and we already have pushed the records with mirrorURLs declaring HTTPS support to the RegTAP service at dc.g-vo.org (the others will have to wait a bit longer, as we haven't re-published our registry records yet (it's all experimental, after all).

![[RSS]](../theme/image/rss.png)