At the Gaia Passivation Event
[All times in CET]
9:00
The instrument that featured most frequently (try this) in this blog is ESA's Gaia Spacecraft that, during the past eleven years, has obtained the positions and much more (my personal favourite: the XP spectra) of about two billion objects, mostly of stars, but also of quasars, asteroids and whatever else is reasonably point-like.
Today, this mission comes to an end. To celebrate it – the mission, not the end, I would say –, ESA has organised a little ceremony at its operations centre in Darmstadt, just next to Heidelberg. To my serious delight, I was invited to that farewell party, and I am now listening to an overview of the passivation given by David Milligan, who used to manage spacecraft operations. This is a suprisingly involved operation, mostly because spacecraft are built to recover from all kinds of mishaps automatically and thus will normally come back on when you switch them off:
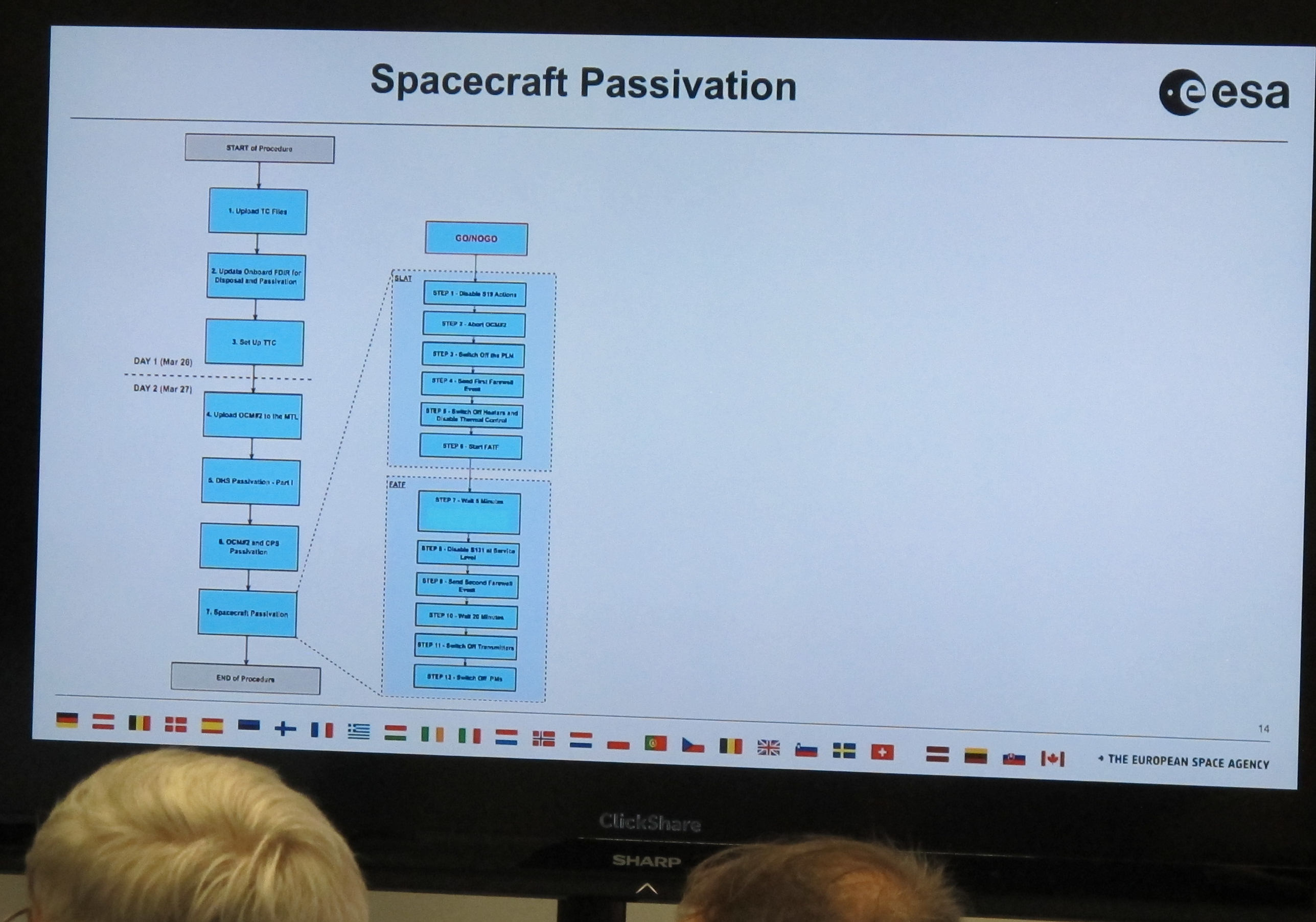
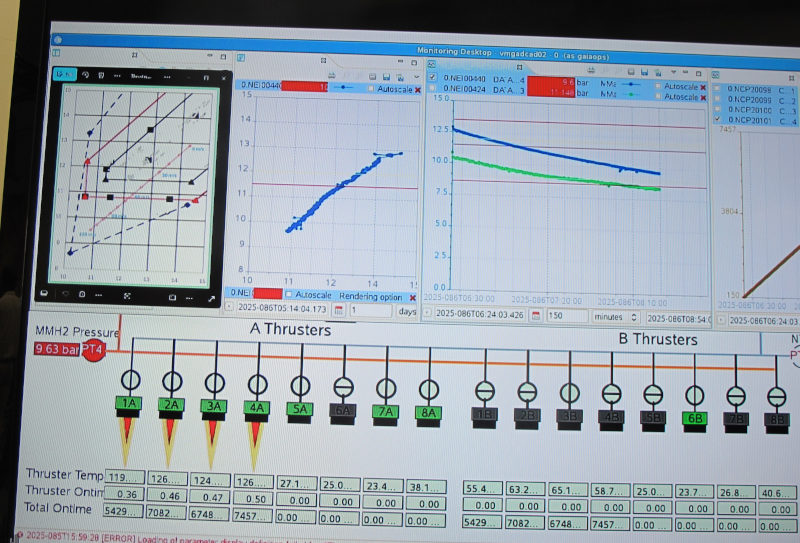
But for now Gaia is still alive and kicking; the control screen shows four thrusters accelerating Gaia out of L2, the Lagrange point behind Earth, where it has been taking data for all these years (if you have doubts, you could check amateur images of Gaia taken while Gaia was particularly bright in the past few months; the service to collect them runs on my data centre).
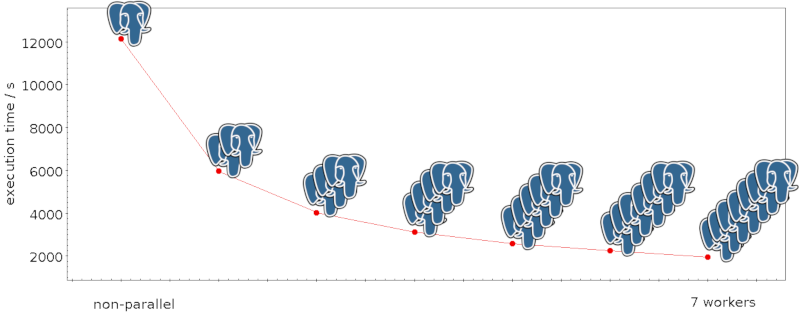
They are working the thrusters quite a bit harder than they were designed for to get to a Δv of 120 m/s (your average race car doesn't make that, but of course it takes a lot less time to accelerate, too). It is not clear yet if they will burn to the end; but even if one fails early, David explains, it is already quite unlikely that Gaia will return.
9:35
Just now the thrusters on the spacecraft have been shut down (”nominally”, as they say here, so they've reached the 120 m/s). Gaia is now on its way into a heliocentric orbit that, as the operations manager said, will bring it back to the Earth-Moon-System with chance of less than 0.25% between now and 2125. That's what much of this is about: You don't want Gaia to crash into anything else that's populating L2 (or something else near Earth, for that matter), or start randomly sending signals that might confuse other missions.
9:42
Gaia is now going blind. They have switched off the science computers a few minutes ago, which we could follow on the telemetry screen, and now they are switching off the CCDs, one after the other. The RP/BP CCDs, the ones that obtained my beloved XP spectra, are already off. Now the astrometry CCDs go grey (on the screen) one after the other. This feels oddly sombre.
In a nerdy joke, they switched off the CCDs so the still active ones formed the word ”bye” for a short moment:
9:45
The geek will inherit the earth. Some nerd has programmed Gaia to send, while it is slowly winding down, an extra text message: “The cosmos is vast. So is our curiosity. Explore!”. Oh wow. Kitsch, sure, but still goosebumpy.
9:50
Another nerdy message: ”Signing off. 2.5B stars. countless mysteries unlocked.” Sigh.
9:53
Gaia is now mute. The operations manager gave a little countdown and then said „mark”. We got to see the spectrum of the signal on the ground station, and then watch it disappear. There was dead silence in the room.
9:55
Gaia was still listening until just now. Then they sent the shutdown command to the onboard computer, so it's deaf, too, or actually braindead. Now there is no way to revive the spacecraft short of flying there. ”This is a very emotional moment,” says someone, and while it sounds like an empty phrase, it is not right now. ”Gaia has changed astronomy forever”. No mistake. And: ”Don't be sad that it's over, be glad that it happened”.
12:00
Before they shut down Gaia, they stored messages from people involved with the mission in the onboard memory – and names of people somehow working on Gaia, too. And oh wow, I found my name in there, too:

It felt a bit odd to have my name stored aboard a spacecraft in an almost eternal heliocentric orbit.
But on reflection: This is solid state storage, in other words, some sort of EPROM. And that means that given the radiation up there, the charges that make up the bit pattern will not live long; colleagues estimated that, with a lot of luck, this might still be readable 20 years from now, but certainly not much longer. So, no, it's not like I now share Kurt Waldheim's privilege of having something of me stored the better part of eternity.
12:30
Andreas Rudolph, head of operations, now gives a talk on, well, ”Gaia Operations”.
I have already heard a few stories of the olden days while chatting to people around here. For instance, ESTEC staff is here and gave some insight views on the stray light trouble that caused quite a few sleepless nights when it was discovered during commissioning. Eventually it turned out it was because of fibres sticking out from the sunshield. Today I learned that had a long history because unfolding the sunshield actually was a hard problem during spacecraft design, and, as Andreas just reminded us, a nail-biting moment during commissioning. The things need to be rollable but stiff, and unroll reliably once in space.
People thought of almost everything. But once they showed the sunshield to an optical engineer while debugging the problem, after a few minutes he shone the flashlight of his telephone behind the screens and conclusively demonstrated the source of the stray light.
Space missions are incredibly hard. Even the smallest oversights can have tremendous consequences (although the mission extension after the original five years of mission time certainly helped offsetting the stray light problem).
Andreas discussed more challenges like that, in particular the still somewhat mysterious Basic Angle Variation, and finished predicting that in 2029, Gaia will next approach Earth, passing at a distance of about 10 million kilometers. I don't think it will be accessible to amateur telescopes, perhaps not even to professional ones. But let's see.
13:00
Gaia data processing is (and will be for another 10 years or so) performed by a large international collaboration called DPAC. DPAC is headed by Anthony Brown, and his is the last talk for today. He mentioned some of the exciting science results of the Gaia mission. Of course, that is a minute sample taken from the thousands and thousands of papers that would not exist without Gaia.
- The tidal bridge of stars between the LMC and the SMC.
- The discovery of 160'000 asteroids (with 5.5 years of data analysed), and their rough spectra, allowing us to group them into several classes.
- The high-precision reference catalogue which is now in use everywhere to astrometrically calibrate astronomical images; a small pre-release of this was already in use for the navigation of the extended mission of the Pluto probe New Horizons.
- Finding the young stars in the (wider) solar neighbourhood by their over-luminosity in the colour-magnitude diagram, which lets you accurately map star forming regions out to a few hundred parsecs.
- Unraveling the history of the Milky Way by reconstructing orbits of hundreds of millions of stars and identifying stellar streams (or rather, overdensities in the space of orbital elements) left over by mergers of other galaxies with the Milky Way and preserved over 10 billion years.
- Confirming that the oldest stars in the Mikly Way are indeed in the bulge using the XP spectra, and reconstructing how the disk formed afterwards.
- In the vertical motions of the disk stars, there is a clear signal of a recent perturbation (probably when the Sagittarius dwarf galaxy crashed through the Milky Way disk) and how there is now some sort of wave going through the disk and slowly petering out.
- Certain white dwarfs (I think those consisting of carbon and nitrogen) show underluminosities because they form bizarre crystals in their outer regions (or so; I didn't quite get that part).
- Thousands of star clusters newly discovered (and a few suspected star clusters debunked). One new discovery was actually hiding behind Sirius; it took space observations and very careful data reduction around bright sources to see it in the vicinity of this source overshining everything around it.
- Quite a few binary stars having neutron stars or black holes as companions – where we are still not sure how some of these systems can even form.
- Acceleration of the solar system: The sun orbits the centre of the Milky Way, once every about 220 Million years or so. So, it does not move linearly, but only very slightly so (“2 Angstrom/s²” acceleration, Anthony put it). Gaia's breathtaking precision let us measure that number for the first time.
- Oh, and in DR4, we will see probably 1000s of new exoplanets in a mass-period range not well sampled so far: Giant planets in wide orbits.
- And in DR5, there will even be limits on low-frequency gravitational waves.
Incidentally, in the question session after Anthony's talk, the grandmaster of both Hipparcos and Gaia, Erik Høg, reminded everyone of the contributions by Russian astronomers to Gaia, among other things having proposed the architecture of the scanning CCDs. I personally have to say that I am delighted to be reminded of how science overcomes the insanities of politics and nationalism.

![[RSS]](../theme/image/rss.png)
![[number line with location markers]](/media/floatingpoint.png)