DaCHS 2.2 is out
DaCHS 2.2 adds support for what simple semantics we currently do in the VO. Which is a welcome excuse to abuse one of the funny symbols semanticians love so much.
Today, I've released DaCHS 2.2, the second stable version of DaCHS running on Python 3. Indeed, we have ironed out a few sore spots that have put that “stable” into question, especially if you didn't run things on Debian Buster. Mind you, playing it safe and just going for Debian is still recommended: Compared to the Python 2 world, where things largely didn't break for a decade, the Python 3 universe is still shaking out, and so the versions of dependencies do matter. It's actually fairly gruesome how badly pyparsing 2.4 will break DaCHS. But that's for another day.
Despite this piece of fearmongering, it'd be great if you could upgrade your installations if you are running DaCHS, and it's pretty safe if you're on Debian buster anyway (and if you're running Debian in the first place, you should be running buster by now).
Here are the more notable changes in this release:
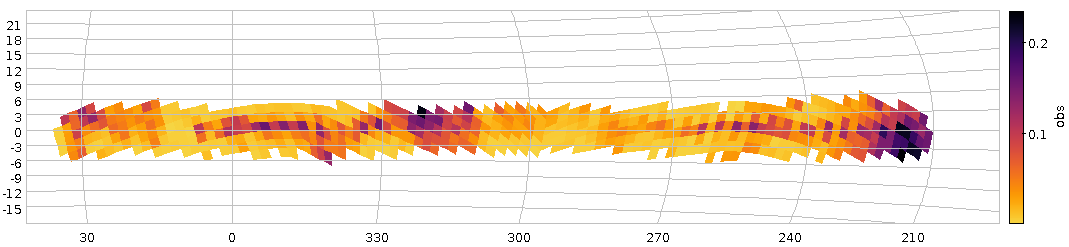
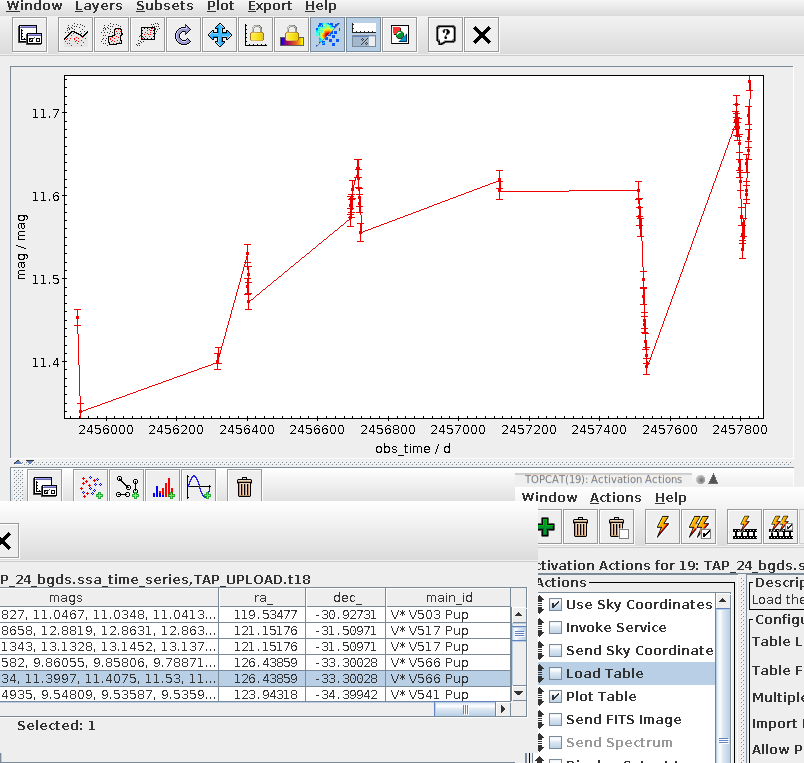
- DaCHS can now (relatively easily) write time series in the form of what Ada Nebot's Time Series Annotation note proposes. See the tutorial chapter on building time series for how to do that in practice. Seriously: If you have time series to publish, by all means try this out. The specification can still be fixed, and so this is the perfect time to find problems with the plan.
- The 2.2 release contains support for the MOC ADQL functions mentioned in the last post on this blog. Of course, to make them work, you will still have to acquaint your database with the new functionality.
- DaCHS has learned to use IVOA vocabularies as per the current draft for Vocabularies in the VO 2. The most visible effect for you probably is that DaCHS now warns if your subject keywords are not taken from the Unified Astronomy Thesaurus (UAT) – which they almost certainly are not, because the actual format of these keywords is a bit funky. On the other hand, if you employ the “plain” root page template (see the root template in our templating guide if you are not sure what I am talking about here), you will get nice, human-friendly labels for the computer-friendly terms you ought to put into subjects. In case you don't bother: Given I'm currently serving as chair of the semantics working group of the IVOA, the whole topic will certainly come up again soon, and at that point I will probably also talk about another semantics-related newcomer to DaCHS, the gavo_vocmatch ADQL UDF.
- There is a new command dachs datapack for interacting with frictionless data packages. The idea is that you can say dachs datapack create myres/q myres.pack and obtain an archive of all that is necessary to re-create myres on another DaCHS installation, where you would say dachs datapack load myres.pack. Frankly, this isn't much different from just tarring up the resource directory at this point, except that any cruft that may have accumulated in the directory is skipped and there is a bit of structured metadata. But then interoperability always starts slowly. Note, by the way, that this certainly does not teach DaCHS to do anything sensible with third-party data packages; while I've not thought hard about this, as it seems a remote use case, I am pretty sure that even the “tabular data packages” that refine the rough general metadata quite a bit simply have nowhere near enough metadata to create a useful VO resource or TAP table.
- As part of my never-ending struggle against bitrot (in case you've always wondered what “curation” means: that, essentially), I'm running dachs val -vc ALL in my own data center once every month. This used to traverse the file system to locate all RDs defined on a box and then make sure they are still ok and their definitions match the database schema. That behaviour has now changed a bit: It will only check published RDs now. I cannot lie: the main reason for the change is because on my production machine the file system traversal has taken longer and longer as data accumulated. But then beyond that there is much less to worry when unpublished gets a little bit mouldy. To get back the old behaviour of validating all RDs that are reachable by the server, use ALL_RECURSE instead of ALL.
- DaCHS has traditionally assumed that you are running multiple services on one site, which is why its root page is rendered over a service that exposes metadata on local resources. If that doesn't quite work for how you use DaCHS – perhaps because you want to have your own custom renderers and data functions on your root page, perhaps because you only have one browser-based service and that should be the root page right away –, you can now override what is shown when people access the root URI of your DaCHS installation by setting the [web]root config item to the path of the resource you want as root (e.g., myres/q/s/fixed when the root page should be made by the fixed renderer on the service s within the RD myres/q).
- Scripting in DaCHS is a powerful way to execute python or SQL code when certain things happen. That seems an odd thing to want until you need it; then you need it badly. Since DaCHS 2.2, scripts executed before or after the creation of a table, before its deletion, or after its meta data has been updated, can sit on tables (where they have always belonged). Before, they could only be on makes (where they can still sit, but of course they are then only executed if the table is operated through that particular make) and RDs (from where they could be copied). That latter location is now forbidden in order to free up RD scripts for later sanitation. Use STREAM and FEED instead if you really used something like that (and I'd bet you don't).
- Minor behavioural changes:
- Due to a bug, you could write things like <schema foo="bar">my_schema<schema>, i.e., have attributes on attributes written in element form. That is now flagged as an error. Since that attribute was fed to the embedding element, you might need to add it there.
- If you have custom flot plots in one of your templates (and you don't if you don't know what I'm talking about), you now have to set style to Points or Lines where you had usingIndex 0 or 1 before.
- The sidebar template no longer has links to a privacy policy (that few bothered to fill out). See extra sidebar items in the tutorial on how to get them back or add something else.
The most important change comes last: The default logo DaCHS shows unless you override it is no longer the GAVO logo. That's, really, been inappropriate from the start. It's now the DaCHS logo, the thing that's in this posts's article image. Which isn't quite as tasteful as the GAVO one, true. But I trust we'll all get used to it.

![[RSS]](../theme/image/rss.png)