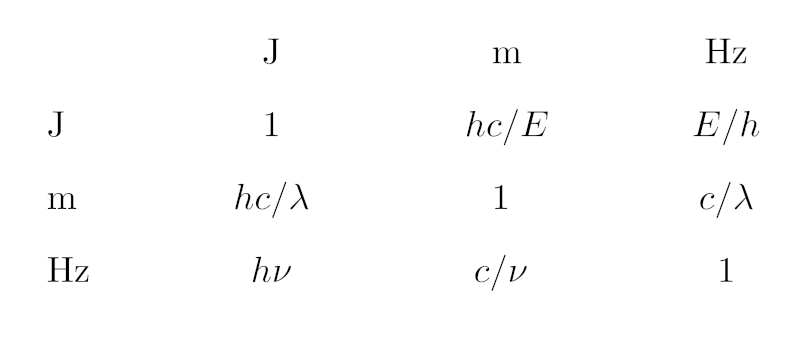GAVO at the Northern Spring Interop 2021
As usual in May, the people making the Virtual Observatory happen meet for their Interoperability Conference, better known as the Interop – where “meet” still has to be taken with a generous helping of salt (more on this near the end of this post). As has become customary on this blog, let me briefly discuss contributions with a significant involvement of GAVO.
A major thing from my perspective actually happened in the run-up: The IVOA executive committee (“Exec“) approved Version 2.0 of Vocabularies in the VO, a standard saying how hierarchical word lists (“vocabularies“) can be managed, disseminated, and consumed within the VO. Developing the main ideas from sufficiently restricting RDF to coming up with desise (which makes complicated things possible with surprisingly little code), and trying things out on our growing number of vocabularies took up quite a bit of my standards time in the last 20 months or so – and I'm fairly happy with the outcome, which I celebrated with a brief talk on programming with IVOA semantics during Wednesday morning's semantics session.
In that session I gave a second, more discussion-oriented, talk, probing how to formalise data product types – which is surprisingly involved, even with the relatively straightforward use case “figure out a programme to handle the data“: What's a spectrum? Well, something that maps a spectral coordinate to... hm. Is it still a spectrum if there's multiple sorts values (perhaps flux, magnitude, and polarisation)? If we allow, in effect, tuples, why not whole images, which would make spectral cubes spectra – but of course few client programmes that deal with spectra do anything useful with cubes, so clearly such a definition would kill our use case. And what about slit spectra, mapping a spatial coordinat to spectra?
All this of course is reminiscent of the classical problems of semantics: An elephant is a big animal with a trunk. But when an elephant loses its trunk in an accident: does it stop being an elephant? So, much of the art here is finding the sweet spot of usability between strict and formal semantics (that will never fit the real world) and just tossing around loosely defined strings (that will simply not be machine-readable). After the session, I came up with the 2021-05-26 draft of product-type. If you read this a few years down the road, it might be interesting to compare with what product-type is today. I'm curious myself.
Later on Wednesday CET, I did a shameless plug for my Datalink-transforming XSLT (apologies for a github link, but I'm fishing for PRs here; if you use DaCHS, you'll get the updated stuff with version 2.4, due soon). The core of this dates back to the dawn of datalink, but with a new graphical cutout code and in particular vocabulary-based tree-ification of the result rows, I figured it's time to remind the operators of datalink services it's still out there for them to take up. Perhaps more than from the slides, you can see what I am after here by just trying the Datalink examples I've collected for this talk and comparing document source, the appearance without Javascript (pure XSLT) and the appearance with Javascript (I'm a bit ashamed I'm relying so heavily on it, but much of this really can only be done client-side).
Quite a bit after midnight my time (still Thursday UTC), Mark Taylor talked about Software Identification, something I've been working on with him recently. It's is one of the things that is short and trivial but that, when unregulated, just doesn't work; in this case it's servers and clients saying what they are when they speak HTTP. I stumbled into the problem while trying to locate severely outdated DaCHS installations – so, I a way I put effort into the Note Mark was talking about (and which I have just uploaded to the IVOA Document Repository) as a sort of penance.
While I was already asleep when Mark gave his talk, I was back at the Interop Friday morning CEST, when Hendrik Heinl talked about the LOFAR TAP service (which, I'm proud to say, runs on top of DaCHS); this was mainly live operations in TOPCAT (which is why there's no exciting slides), but Hendrik used a pyVO script doing cutouts in an (optical) mosaic of the Fornax cluster built on top of – and that's the main point – Datalink and SODA. Working this out with Hendrik made me realise the documentation of Datalink in pyVO really needs… love. Or, better, work.
Later on Friday, there was the Registry session, where I gave brief (and somewhat cramped) talks on advanced column metadata (which is intended to one day let you query the registry for things like “roughly complete to 18 mag” or “having objects out to redshift 4“) and how to put VODataService 1.2 coverage into RegTAP – I expect you'll read more on both topics on this blog as they mature to a level at which this can leave the Registry nerd circles.
And now, about 10 pm on Friday, the meeting is slowly winding down; beyond all the talks (which were, regrettably for a free software spirit like me, on zoom), the real bonus was that there was a gather.town attached to the conference. Now, that's a closed, proprietary, non-self-hostable platform, too, and so I have all reason to grumble. But: for the first time since February 2020 it felt like a conference, with the most useful action happening outside of the lecture halls, from trying to reach consensus on VEP-006 to teaching DaCHS datalink service declaration to learning about working with visibilities coming from VLBI (where it's even more difficult than it is with the big antenna arrays). So… this one time I've made my peace with proprietary platforms.
A propos of “say no to platforms“ (in this case, slack): Due to the recent troubles with freenode, in addition to the Interop last week saw the the GAVO IRC channel move to libera.chat (where it's still #gavo). So, for instant messaging us now that the Interop is (in effect) over: Come there.

![[RSS]](./theme/image/rss.png)