The 2021 Southern Spring Interop
A contribution for the ”things that didn't work out” (“Arbeiten, die zu keiner Lösung geführt haben”) section in our reports to BMBF: an attempt to systematise product types at the last Interop. I've made a new proposal at this Interop, and there is reason to hope it will fare better.
Last night, the second IVOA Interop conference of 2021 came to an end; I'm calling it ”southern spring” because notionally, it happened in Cape Town, back to back with this year's ADASS. In reality, it was again an online event, and so, in keeping up with the tradition established in the pandemic times, the closing event was around midnight CET. I cannot say I will miss these late-night events, although I would not go as far as some people at the conference who quipped they'd prefer the airport security checks to having to sit through another zoom marathon.
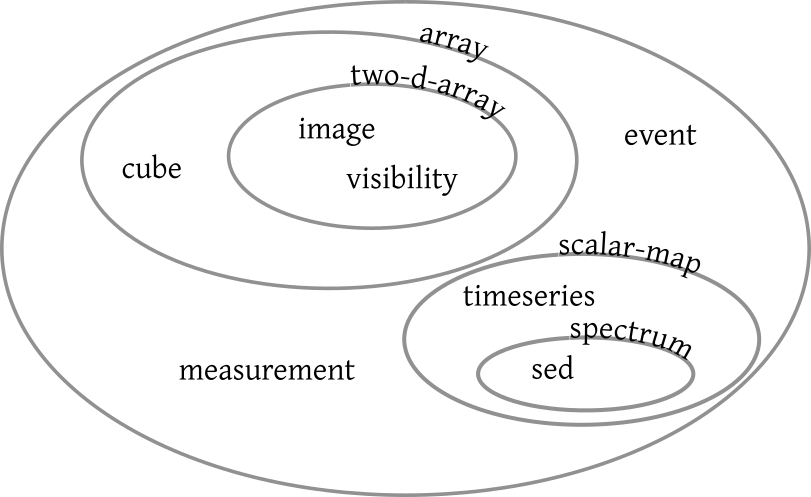
My contributions at this interop again had a clear focus on semantics, for instance with my public confession that my attempt to systematise “product types” at the last interop was entirely misguided; trying to force concepts like “time series”, “spectrum“ or “image” into a tree does not lead to anything that actually works for what this is intended to do, that is, helping people find the sort of data they are after for a particular purpose, or helping clients route data products to other clients better suited to process them. I will now try a restart using SKOS, a plan that was met with a lot more agreement than that previous attempt. Some entertainment at the side was provided by the realisation that a “time-image cube“ is normally called a movie. Next time I'll take in moving pictures, I'll find out what people say when I claim to investigate a time cube.
Another talk that took up a topic from the last Interop's Semantics session was about making an IVOA vocabulary of object types based on the work done within the CDS over the last 40 year or so. This certainly is just the beginning of a longer effort, not the least because the current concepts severely fall short in the area of the solar system. But it's a start, and there's plenty of time to elaborate this before it will go through a review, presumably with the next version of Obscore.
Also semantics-related, but over in the session of the Operations interest group, Mark Taylor reported on his activities to evaluate the standards adherence of semantics information in published tables. This activity is what had triggered me to make DaCHS validate UCDs assigned to columns in summer, something that I expect will result in quite few diagnostics when DaCHS operators upgrade to DaCHS 2.5 (expected for November). But that's fine: making it more likely that computers will actually recognise a, say, error in proper motion for what it is is undoubtedly a good thing. I'm therefore glad that there is almost a million “good” UCDs out there and a lot fewer somehow “bad”. I had expected much worse after my realisation that my own annotations left a lot to be desired in summer. By now, the only bad UCDs I'm still pushing out are the ones mandated by SSAP and SLAP. The contradictions between those standards and UCD are going to be addressed with Errata in the coming months.
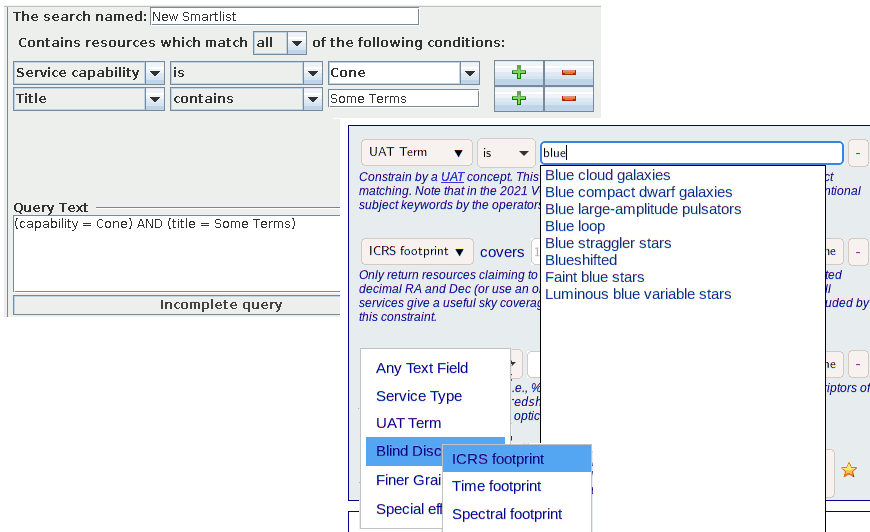
My talk in the third Apps session on Thursday afternoon still had some relationship with Semantics; it was a quick show and tell on the enhancements to WIRR I had reported on here in July, and it in particular showcased obtaining UCD constraints by full-text searching the rr.table_column table in my RegTAP service and the selection through UAT concepts. Satisfyingly in some way, it were these topics that people took up in the discussion after the talk. Less satisfyingly, people playing with the thing afterwards turned up something that has the alarming taste of a bug in the new MOC operations in pgsphere. Ouch.
This segues into the realm of Registry, where there was no actual session but a rather well-attended side meeting in the gathertown instance we could take over from ADASS (that, incidentally, was substantially better attended than during the previous meetings). There, I mainly presented (and explained) my proposed changes to pyVO's registry interface currently living in a private branch in my fork on github. I will write a bit more on that around the time I will turn that into a PR.
Another outcome of this was that there was some interest to turn the note on documents in the Registry – which is what feeds VOTT – into either an endorsed note or perhaps a Recommendation of the Registry WG.
My fourth “proper” (in the rather twisted sense of: in a zoom session) talk was an attempt to finally do something about the problems pointed out in my caproles note lamenting that our current service registration patterns are fundamentally flawed. It proposed some ways to to get VOSI availability fixed, and the outcome was that we probably will drop what we currently require in that field, not the least because these requirements are cheerily ignored by 98% of the resources in the Registry.
Those were again three fairly long days, usually starting with sessions around 7:00 CET and ending with sessions around midnight. Which is clearly not healthy. But on the other hand, it somehow does convey a physical sense of the global nature of the Virtual Observatory, on which people in many, many time zones work. And that, I have to say, still is something I do appreciate.

![[RSS]](./theme/image/rss.png)