2020-09-22
Markus Demleitner
So far when you did spherical geometry in ADQL, you had points, circles,
and polygons as data types, and you could test for intersection and
containment as operations. This feature set is a bit unsatisfying
because there are no (algebraic) groups in this picture: When you join
or intersect two circles, the result only is a circle if one contains
the other. With non-intersecting polygons, you will again not have a
(simply connected) spherical polygon in the end.
Enter MOCs (which I've
mentioned a few times before on this
blog): these are essentially arbitrary shapes on the sky, in practice
represented through lists of pixels, cleverly done so they can be
sufficiently precise and rather compact at the same time. While MOCs are
powerful and surprisingly simple in practice, ADQL doesn't know about
them so far, which limits quite a bit what you can do with them. Well,
DaCHS would serve them since about 1.3 if you managed to push them into
the database, but there were no operations you could do on them.
Thanks to work done by credativ (who were
really nice to work with), funded with some money we had left from our
previous e-inf-astro project (BMBF FKZ 05A17VH2) on the pgsphere
database extension, this has
now changed. At least on the GAVO data center, MOCs are now essentially
first-class citizens that you can create, join, and intersect within
ADQL, and you can retrieve the results. All operators of DaCHS services
are just a few updates away from being able to offer
the same.
So, what can you do? To follow what's below, get a sufficiently new
TOPCAT (4.7 will do) and open its TAP client on http://dc.g-vo.org/tap
(a.k.a. GAVO DC TAP).
Basic MOC Operations in TAP
First, let's make sure you can plot MOCs; run
SELECT name, deepest_shape
FROM openngc.shapes
Then do Graphics/Sky Plot, and in the window that pops up then,
Layers/Add Area Control. Then select your new table in the Position tab,
and finally choose deepest_shape as area (yeah, this could become a bit
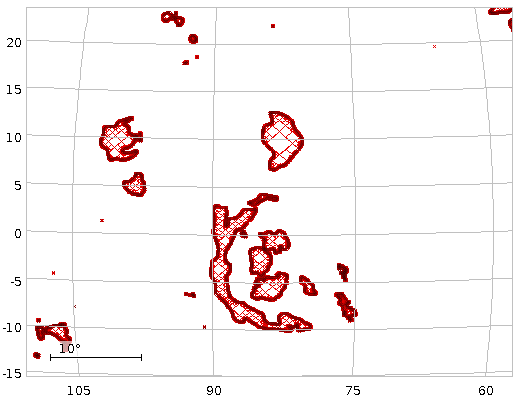
more automatic and probably will over time). You will then see the
footprints of a few NGC objects (OpenNGC's author Mattia Verga hasn't
done all yet; he certainly welcomes help on OpenNGC's version control
repo), and you can move
around in the plot, yielding perhaps something like Fig. 1.
Now let's color these shapes by object class. If you look, openngc.data
has an obj_type column – let's group on it:
SELECT
obj_type,
shape,
AREA(shape) AS ar
FROM (
SELECT obj_type, SUM(deepest_shape) AS shape
FROM openngc.shapes
NATURAL JOIN openngc.data
GROUP BY obj_type) AS q
(the extra subquery is a workaround necessary because the area
function wants a geometry or a column reference, and ADQL doesn't allow
aggregate functions – like sum – as either of these).
In the result you will see that so far, contours for about 40 square
degrees of star clusters with nebulae have been put in, but only 0.003
square degrees of stellar associations. And you can now plot by the
areas covered by the various sorts of objects; in Fig. 2, I've used
Subsets/Classify by Column in TOPCAT's Row Subsets to have colours
indicate the different object types – a great workaround when one deals
with categorial variables in TOPCAT.
MOCs and JOINs
Another table that already has MOCs in them is rr.stc_spatial, which has
the coverage of VO resources (and is the deeper reason I've been pushing
improved MOC support in pgsphere – background); this isn't available
for all resources yet , but at least there are about 16000 in already.
For instance, here's how to get the coverage of resources talking about
planetary nebulae:
SELECT ivoid, res_title, coverage
FROM rr.subject_uat
NATURAL JOIN rr.stc_spatial
NATURAL JOIN rr.resource
WHERE uat_concept='planetary-nebulae'
AND AREA(coverage)<20
(the rr.subject_uat table is a local extension to RegTAP that will be
the subject of some future blog post; you could also use rr.res_subject,
but because people still use wildly different keyword schemes – if any
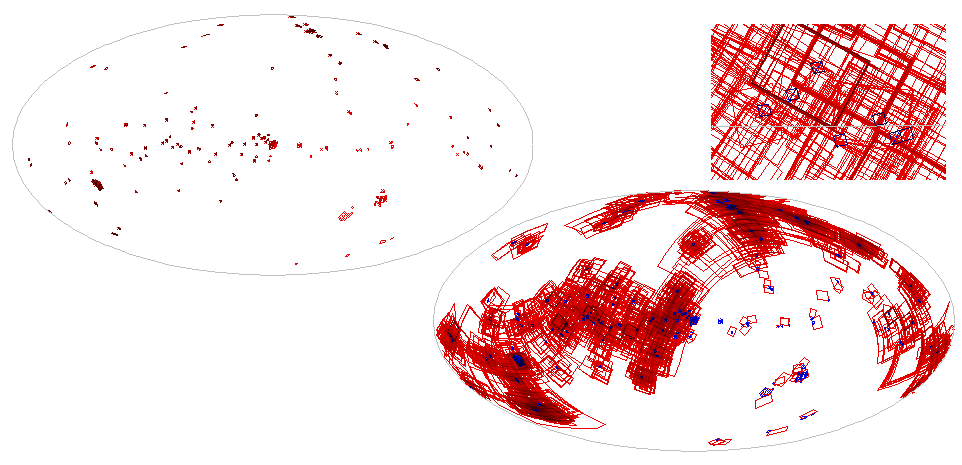
–, that wouldn't be as much fun). When plotted, that's the left side of
Fig. 3. If you do that yourself, you will notice that the resolution
here is about one degree, which is a special property of the sort of
MOCs I am proposing for the Registry: They are of order 6. Resolution in
MOC goes up with order, doubling with every step. Thus MOCs of order 7
have a resolution of about half a degree, MOCs of order 5 a resolution
of about two degrees.
One possible next step is fetch the intersection of each of these
coverages with, say, the DFBS (cf. the post on Byurakan spectra). That
would look like this:
SELECT
ivoid,
res_title,
gavo_mocintersect(coverage, dfbscoverage) as ovrlp
FROM (
SELECT ivoid, res_title, coverage
FROM rr.subject_uat
NATURAL JOIN rr.stc_spatial
NATURAL JOIN rr.resource
WHERE uat_concept='planetary-nebulae'
AND AREA(coverage)<20) AS others
CROSS JOIN (
SELECT coverage AS dfbscoverage
FROM rr.stc_spatial
WHERE ivoid='ivo://org.gavo.dc/dfbsspec/q/spectra') AS dfbs
(the DFBS' identifier I got with a quick query on WIRR). This uses the
gavo_mocintersect user defined function (UDF), which takes two MOCs
and returns a MOC of their common pixels. Which is another important
part why MOCs are so cool: together with union and intersection, they
form groups. It should not come as a surprise that there is also a
gavo_mocunion UDF. The sum aggregate function we've used in our
grouping above is (conceptually) built on that.
You can also convert polygons and circles to MOCs using the (still
DaCHS-only) MOC constructor. For instance, you could compute the
coverage of all resources dealing with planetary nebulae, filtering
against obviously over-eager ones by limiting the total area, and then
match that against the coverages of images in, say, the Königstuhl plate
achives HDAP. Watch this:
SELECT
im.*,
gavo_mocintersect(MOC(6, im.coverage), pn_coverage) as ovrlp
FROM (
SELECT SUM(coverage) AS pn_coverage
FROM rr.subject_uat
NATURAL JOIN rr.stc_spatial
WHERE uat_concept='planetary-nebulae'
AND AREA(coverage)<20) AS c
JOIN lsw.plates AS im
ON 1=INTERSECTS(pn_coverage, MOC(6, coverage))
– so, the MOC(order, geo) function should give you a MOC for other
geometries. There are limits to this right now because of limitations of
the underlying MOC library; in particular, non-convex polygons are not
supported right now, and there are precision issue. We hope this will be
rectified soon-ish when we base pgsphere's MOC operations on the CDS
HEALPix library.
Anyway, the result of this is plotted on the right of Fig. 3.
Open Ends
In case you have MOCs from the outside, you can also construct MOCs from
literals, which happen to be the ASCII MOCs from the standard. This could look like this:
SELECT TOP 1
MOC('4/30-33 38 52 7/324-934') AS ar
FROM tap_schema.tables
For now, you cannot combine MOCs in CONTAINS and INTERSECTS expressions
directly; this is mainly because in such an operation, the machine as to
decide on the order of the MOC the other geometries are converted to
(and computing the predicates between geometry and MOC directly is
really painful). This means that if you have a local table with MOCs in
a column cmoc that you want to compare against a polygon-valued
column coverage in a remote table like this:
SELECT db.* FROM
lsw.plates AS db
JOIN tap_upload.t6
ON 1=CONTAINS(coverage, cmoc) -- fails!
you will receive a rather scary message of the type “operator does not
exist: spoly <@ smoc”. To fix it (until we've worked out how to
reasonably let the computer do that), explicitly convert the polygon:
SELECT db.* FROM
lsw.plates AS db
JOIN tap_upload.t6
ON 1=CONTAINS(MOC(7, coverage), cmoc)
(be stingy when choosing the order here – MOCs that already exist are
fast, but making them at high order is expensive).
Having said all that: what I've written here is bleeding-edge, and it is
not standardised yet. I'd wager, though, that we will see MOCs in ADQL
relatively soon, and that what we will see will not be too far from this
experiment. Well: Some rough edges, I'd hope, will still be smoothed
out.
Getting This on Your Own DaCHS Installation
If you are running a DaCHS installation, you can contribute to takeup
(and if not, you can stop reading here). To do that, you need to upgrade
to DaCHS's latest beta (anything newer than 2.1.4 will do) to have the
ADQL extension, and, even more importantly, you need to install the
postgresql-postgres package from our release repository (that's version
1.1.4 or newer; in a few weeks, getting it from Debian testing would
work as well).
You will probably not get that automatically, because if you followed
our normal installation instructions, you will have a package called
postgresql-11-pgsphere installed (apologies for this chaos; as ususal,
every single step made sense). The upshot is that with our release repo
added, sudo apt install postgresql-pgsphere should give you the new
code.
That's not quite enough, though, because you also need to acquaint the
database with the new functions. This can only be done with database
administrator privileges, which DaCHS by design does not possess. What
DaCHS can do is figure out the commands to do that when it is called as
dachs upgrade -e. Have a look at the output, and if you are
satisfied it is about what to expect, just pipe it into psql as a
superuser; in the default installation, dachsroot would be sufficiently
privileged. That is:
dachs upgrade -e | psql gavo # as dachsroot
If running:
select top 1 gavo_mocunion(moc('1/3'), moc('2/9'))
from tap_schema.tables
through your TAP endpoint returns '1/3 2/9', then all is fine. For
entertainment, you might also make sure that
gavo_mocintersect(moc('1/3'), moc('2/13')) is 2/13 as expected, and
that if you intersect with 2/3 you get back an empty string.
So – let's bring MOCs to ADQL!
![[number line with location markers]](/media/floatingpoint.png)

![[RSS]](../theme/image/rss.png)